Abstract
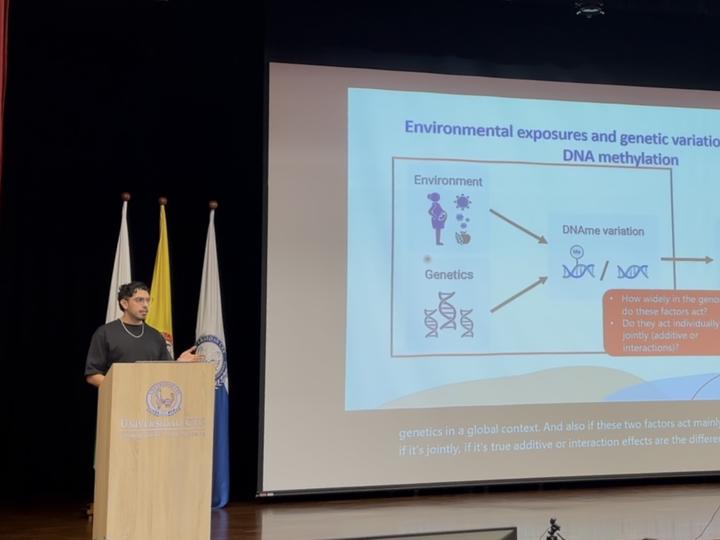
DNA methylation (DNAme) is an epigenetic mark that has gained attention under the Developmental Origins of Health and Disease framework due to its association with phenotypic changes and potential long-term stability. Genomic (G) and exposomic (E; totality of exposures a person experiences) differences have been reported as main sources of inter-individual DNAme variability. However, further research is needed to understand the genomic location and characteristics of regions with variable DNAme, the degree to which genetics and environmental factors contribute to DNAme variability in these regions, and whether genetics and environment associate individually with DNAme or in combination. Here, we estimate the genome-exposome contribution to cord blood methylome variability in an integrative multi-omics analysis. Method: We identify genome-wide cord blood Variable Methylated Regions in two independent cohorts (CHILD and PREDO; overall n=1,662) accounting for the sparse and non-homogeneous coverage in the Illumina EPIC v1 array. To reduce the universe of compared models, we implement VMR-wise E and G variable selection procedure based on LASSO. We then fit single-variable G, E, pairwise additive (G+E) and interaction (GxE) linear models of VMR’s DNAme, and select the best one per VMR based on their AIC. To control for spurious associations, we implement a computationally fast permutation approach. Results: With a high percentage of winner models (59%), a higher proportion of DNAme variance explained (M = 0.22), and a full consistency across cohorts (100%), we identify genetics as a consistent main contributor to DNAme variability in cord blood usually in additive and interaction combinations with the prenatal exposome (44.4%). These results showcase the relevance of genetics in diverging DNAme patterns in early life. Furthermore, we introduce RAMEN (Regional Association of Methylome variability with Exposone and geNome; github.com/ErickNavarroD/RAMEN), a user-friendly R package for DNAme microarrays that improves the extraction of regional DNAme variability patterns and models their contributors while controlling for spurious associations expected by chance through state-of-the-art statistical techniques.